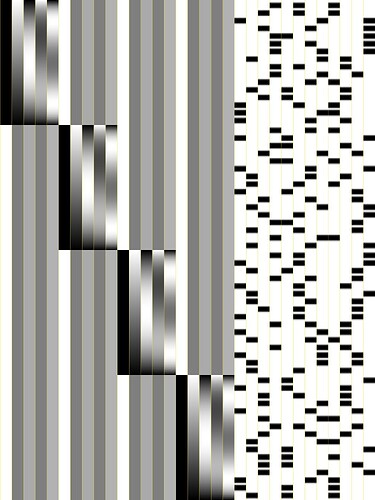
Hi all-
I'm modifying a task and trying to evaluate the design using 3dDeconvolve -nodata.
Each block of the task consists of a combination of three different conditions and there are four variants of each condition (64 possible combinations, each possible combination appears once). I'd like to run a model looking at each of the 12 possible variants (each variant appears 16 times). Given the complexity of the task there is bound to be some degree of collinearity, but I'm wondering if there is too much collinearity.
I get "++ VERY GOOD ++" for all of the matrix conditions, but I am seeing a "** BEWARE **" Matrix inverse average error and very large norm. std. dev. values (in the thousands [see below]). I've played around with some of the task parameters, increasing the number of runs, varying the ITIs, etc., but I still get similar results. Is this design/analysis plan acceptable? How concerned should I be with the matrix inverse average error and norm std dev values?
3dDeconvolve command
3dDeconvolve -nodata 1840 1 \
-concat '1D: 0 460 920 1380' \
-num_stimts 12 \
-stim_times 1 all_both.1D 'BLOCK(20,1)' \
-stim_times 2 all_notboth.1D 'BLOCK(20,1)' \
-stim_times 3 all_either.1D 'BLOCK(20,1)' \
-stim_times 4 all_neither.1D 'BLOCK(20,1)' \
-stim_times 5 all_red.1D 'BLOCK(20,1)' \
-stim_times 6 all_vert.1D 'BLOCK(20,1)' \
-stim_times 7 all_high.1D 'BLOCK(20,1)' \
-stim_times 8 all_constant.1D 'BLOCK(20,1)' \
-stim_times 9 all_li.1D 'BLOCK(20,1)' \
-stim_times 10 all_lm.1D 'BLOCK(20,1)' \
-stim_times 11 all_ri.1D 'BLOCK(20,1)' \
-stim_times 12 all_rm.1D 'BLOCK(20,1)' \
-xjpeg nodata -polort A
The output
*+ WARNING: no -stim_label given for stim #1 ==> label = 'Stim#1'
*+ WARNING: no -stim_label given for stim #2 ==> label = 'Stim#2'
*+ WARNING: no -stim_label given for stim #3 ==> label = 'Stim#3'
*+ WARNING: no -stim_label given for stim #4 ==> label = 'Stim#4'
*+ WARNING: no -stim_label given for stim #5 ==> label = 'Stim#5'
*+ WARNING: no -stim_label given for stim #6 ==> label = 'Stim#6'
*+ WARNING: no -stim_label given for stim #7 ==> label = 'Stim#7'
*+ WARNING: no -stim_label given for stim #8 ==> label = 'Stim#8'
*+ WARNING: no -stim_label given for stim #9 ==> label = 'Stim#9'
*+ WARNING: no -stim_label given for stim #10 ==> label = 'Stim#10'
*+ WARNING: no -stim_label given for stim #11 ==> label = 'Stim#11'
*+ WARNING: no -stim_label given for stim #12 ==> label = 'Stim#12'
++ 3dDeconvolve: AFNI version=AFNI_24.2.01 (Jul 16 2024) [64-bit]
++ Authored by: B. Douglas Ward, et al.
++ using TR=1 seconds for -stim_times and -nodata
++ using NT=1840 time points for -nodata
++ Imaging duration=460.0 s; Automatic polort=4
++ -stim_times using TR=1 s for stimulus timing conversion
++ -stim_times using TR=1 s for any -iresp output datasets
++ [you can alter the -iresp TR via the -TR_times option]
++ ** -stim_times NOTE ** guessing GLOBAL times if 1 time per line; LOCAL otherwise
++ ** GUESSED ** -stim_times 1 using LOCAL times
++ ** GUESSED ** -stim_times 2 using LOCAL times
++ ** GUESSED ** -stim_times 3 using LOCAL times
++ ** GUESSED ** -stim_times 4 using LOCAL times
++ ** GUESSED ** -stim_times 5 using LOCAL times
++ ** GUESSED ** -stim_times 6 using LOCAL times
++ ** GUESSED ** -stim_times 7 using LOCAL times
++ ** GUESSED ** -stim_times 8 using LOCAL times
++ ** GUESSED ** -stim_times 9 using LOCAL times
++ ** GUESSED ** -stim_times 10 using LOCAL times
++ ** GUESSED ** -stim_times 11 using LOCAL times
++ ** GUESSED ** -stim_times 12 using LOCAL times
++ Number of time points: 1840 (no censoring)
- Number of parameters: 32 [20 baseline ; 12 signal]
++ Wrote matrix image to file nodata.jpg
++ Wrote matrix values to file nodata.xmat.1D
++ ----- Signal+Baseline matrix condition (1840x32): 2.21842 ++ VERY GOOD ++
*+ WARNING: !! in Signal+Baseline matrix:
- Largest singular value=1.96037
- 2 singular values are less than cutoff=1.96037e-07
- Implies strong collinearity in the matrix columns!
++ Signal+Baseline matrix singular values:
0 0 0.398334 0.562453 0.605234
0.667968 0.716197 0.728911 0.763709 0.810864
0.845201 0.863785 0.993715 0.994667 0.996958
0.998195 0.998801 1.00117 1.00224 1.00426
1.00494 1.00624 1.12005 1.13376 1.15804
1.19266 1.21188 1.22015 1.24485 1.27737
1.29586 1.96037
++ ----- Signal-only matrix condition (1840x12): 1.31652 ++ VERY GOOD ++
*+ WARNING: !! in Signal-only matrix:- Largest singular value=1.73267
- 2 singular values are less than cutoff=1.73267e-07
- Implies strong collinearity in the matrix columns!
++ Signal-only matrix singular values:
0 0 0.999676 0.99976 0.999793
0.999853 0.999874 0.999897 0.999999 1.00001
1.00006 1.73267
++ ----- Baseline-only matrix condition (1840x20): 1.00728 ++ VERY GOOD ++
++ ----- polort-only matrix condition (1840x20): 1.00728 ++ VERY GOOD ++
++ Wrote matrix values to file nodata_XtXinv.xmat.1D
*+ WARNING: +++++ !! Matrix inverse average error = 0.0153459 ** BEWARE **
++ Matrix setup time = 3.47 sStimulus: Stim#1
h[ 0] norm. std. dev. = 172503.7031Stimulus: Stim#2
h[ 0] norm. std. dev. = 172503.7031Stimulus: Stim#3
h[ 0] norm. std. dev. = 172503.7031Stimulus: Stim#4
h[ 0] norm. std. dev. = 172503.7031Stimulus: Stim#5
h[ 0] norm. std. dev. = 156609.9844Stimulus: Stim#6
h[ 0] norm. std. dev. = 156609.9844Stimulus: Stim#7
h[ 0] norm. std. dev. = 156609.9844Stimulus: Stim#8
h[ 0] norm. std. dev. = 156609.9844Stimulus: Stim#9
h[ 0] norm. std. dev. = 164366.0312Stimulus: Stim#10
h[ 0] norm. std. dev. = 164366.0312Stimulus: Stim#11
h[ 0] norm. std. dev. = 164366.0312Stimulus: Stim#12
h[ 0] norm. std. dev. = 164366.0312
I'd be happy to share the stimulus timing files if that would be helpful.
Thanks
AFNI version info (afni -ver): Precompiled binary macos_10.12_local: Jul 16 2024 (Version AFNI_24.2.01 'Macrinus')