AFNI version info (afni -ver): Precompiled binary linux_openmp_64: Jun 10 2021 (Version AFNI_21.1.10 'Domitian')
Code run:
3dTcat -prefix modgroupxtarg_concat.nii fl.modgroupxtarg.pvalue.nii fl.modgroupxtarg.fvalue.nii
3dClusterize -inset modgroupxtarg_concat.nii -ithr 1 -idat 0 -mask harmonized_groupmask.nii -NN 1 -1sided 'RIGHT' 5 -clust_nvox 36 -pref_map ClusterMap_1sided_5 > ClusterReport_1sided_5thr
Hi,
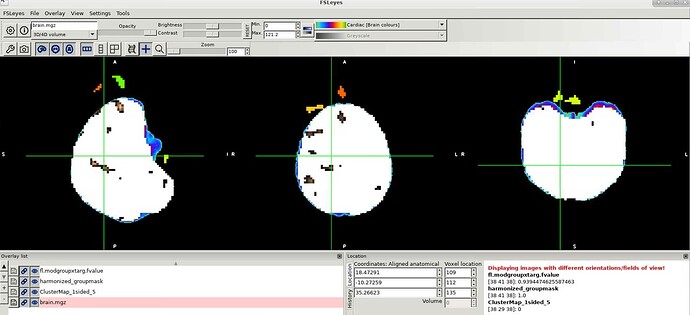
I'm running 3dClusterize on stats output I have from another program. I've combined the f-stat and p-value maps into one, and am running 3dClusterize on that with our cluster size from 3dClustSim. The output looks like I'd expect...except that it seems to be shifted, in a different space than the input. Both input files (mask=white in the image below, brown=modgroupxtarg_concat.nii) are in the same space (mni152 brain.mgz=blue outline underneath image), but the resulting cluster map from 3dClsuterize (clustermap=colored clusters) appear anteriorly shifted from the inputs. Any idea why this might be
happening?
Hmm. The first thing I would check is that modgroupxtarg_concat.nii, ClusterMap_1sided_5, fl.modgroupxtarg.pvalue.nii and fl.modgroupxtarg.fvalue.nii are on the same grid, which I presume they should be:
3dinfo -same_all_grid \
modgroupxtarg_concat.nii \
ClusterMap_1sided_5+tlrc \
fl.modgroupxtarg.pvalue.nii \
fl.modgroupxtarg.fvalue.nii
If all of those are on the same grid, you should get four rows of five 1s.
You could convert the mni152 brain.mgz to NIFTI or BRIK/HEAD and also check that, too. Note that datasets can be in the same "space" but on different "grids", but the 3dinfo command will tell you what, if any, properties differ by which of the five items checked per dataset produce a 0 (see the help file for 3dinfo about the order of checks).
--pt
3dinfo -same_all_grid modgroupxtarg_concat.nii ClusterMap_1sided_5+orig fl.modgroupxtarg.pvalue.nii fl.modgroupxtarg.fvalue.nii
1 1 1 1 1
1 1 1 1 1
1 1 1 1 1
1 1 1 1 1
I'll note that my 3dClusterize output is ClusterMap_1sided_5+orig, not ClusterMap_1sided_5+tlrc
Also, when I look at the brain.mgz, it appears to not be aligned with the others in the same grid
3dinfo -same_all_grid modgroupxtarg_concat.nii ClusterMap_1sided_5+orig fl.modgroupxtarg.pvalue.nii fl.modgroupxtarg.fvalue.nii brain.nii
1 1 1 1 1
1 1 1 1 1
1 1 1 1 1
1 1 1 1 1
0 0 0 0 1
--athena
Hi, Athena-
Thanks for sending that. From the 2nd command there, last line, indeed, brain.mgz has different:
matrix dimensions, voxel size, header orientation and geometric center. The only thing that matches is the obliquity (which I would wager is zero).
Now, something to consider: things can be in the same space but on different grids. In that case, overlaying the datasets in the AFNI GUI would show them appropriately where they are (because it uses the xyz coords to place things and overlay them). Some other GUIs only use matrix size and things like that, so they might appear off. From the image you showed above, I don't think you were visualizing results in the AFNI GUI.
For doing calculations with datasets, you would indeed want all 5 properties to match and be on the same grid, regardless of software.
I'm guessing from the name+extension "brain.mgz" that that dataset is output by FreeSurfer's recon-all. Note that that program will by default resample every output to 1x1x1 mm^3 voxels on a 256x256x256 matrix grid, so I wouldn't be surprised if its output grid is different than what was input. That isn't necessarily a problem, just something to be aware of.
--pt
Oh, and if you are sure that the datasets are on different grids but really in the same space, you could try resampling the brain.mgz dataset to match with the grid of one of your other datasets of interest. I think you would first have to convert it to either NIFTI or BRIK/HEAD (in AFNI-land, we run @SUMA_Make_Spec_FS on recon-all outputs to convert the volumetric data to NIFTI and surface data to GII and NIML.DSET, to be used AFNI+SUMA). Then, you could run:
3dresample \
-input DSET_BRAIN_MASK \
-master modgroupxtarg_concat.nii \
-prefix DSET_BRAIN_MASK_RES \
-rmode NN
... where the "master" dataset provides the grid properties that the output should have, and the rmode NN is appropriate for an integer-valued dataset that should stay integer valued (it means that the resampling mode is "nearest neighbor", to preserve local values without interpolation).
--pt
Excellent, thank you for the explanation. I'll try that!